Breakthrough in Protein Loop Modeling with Deep Learning Methods
A transformative study led by Professor Tingjun Hou at Zhejiang University introduces a novel deep learning approach for protein loop modeling, published under the title "Highly accurate and efficient deep learning paradigm for full-atom protein loop modeling with KarmaLoop" in the journal Research (DOI: 10.34133/research.0408). This groundbreaking work enhances our ability to predict protein structures with unmatched precision, particularly in the highly variable loop regions that are crucial for biological functions such as enzyme regulation and molecular interactions.
Significance of the Research: Protein loops, which connect secondary structures of protein, present significant challenges in structural prediction due to their flexibility and variability. Accurate modeling of these loops is critical as errors can lead to incorrect predictions of protein behavior, impacting drug design and disease understanding. KarmaLoop addresses these challenges by providing a robust deep-learning method that can adopt high accuracy and efficiency. The innovations include: (1) Enhanced protein representation by merging atom features into residues; (2) Improved accuracy using a pre-trained MDN block to better predict atom distances; (3) Increased efficiency with EGNN-predicted pseudo forces updating loop conformations.
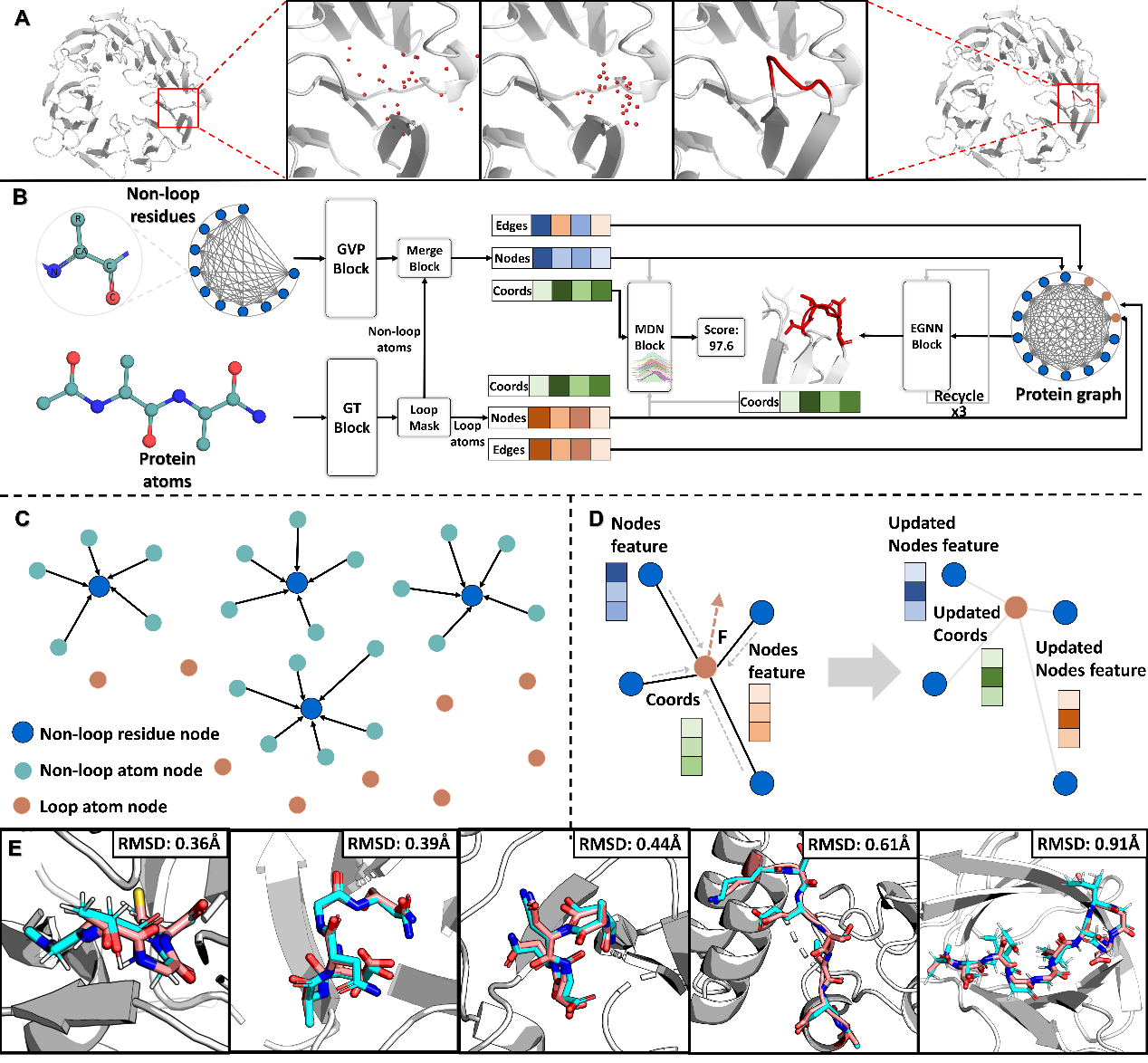
Figure Workflow of KarmaLoop for Loop Modeling. (A) Example of loop modeling, showing the transition from a protein with missing loop regions to a complete protein with predicted loop coordinates. (B) KarmaLoop architecture. (C) Process of hierarchical representation of protein. (D) Feature and coordinate updating process for nodes. (E) Examples of predictions made by KarmaLoop.
Impact and Applications: The accurate prediction of protein loops by KarmaLoop marks a significant advancement in computational biology, offering profound implications for medical research, pharmaceutical developments, and the broader understanding of protein functionality. This study advances protein structure prediction, particularly by enhancing the accuracy of loop modeling, and offers new tools for addressing complex biological modeling challenges.
Tag: Health Science
Source: https://spj.science.org/doi/10.34133/research.0408
